MsCompare for Peak Matching & Peak Picking in Metabolomics
The MsCompare Module is specialized in handling series of samples from Metabolomics and BioMarker Discovery projects. The module’s engine contains two very fast high resolution (or nominal) algorithms to find all the Peaks in all Samples:
-
Peak Matching
-
Peak Picking
Peak Matching, as opposed to Peak Picking, uses one set of reference ions from a single sample. This set of ions will be used to calculate results for a large group of samples with or without alignment. A good procedure to create a proper reference sample is to pool equal aliquots from all samples. These samples are optimal for Quality Control and for Peak Picking as all ions from all samples will be present.
Peak Picking will detect all peaks from all samples independently and combine results using a clustering algorithm. Peak Picking at Peak Matching can be run at any resolution and is also available for GC/MS Data Processing.
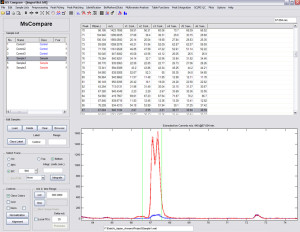
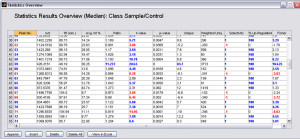
After finding all the peaks, you can perform both univariate and multivariate analysis to find peaks that are significantly different between the groups or classes.
- Univariate Statistics: t-values, uniqueness, ratio, fisher discriminant score etc.
- Quickly remove all non-relevant peaks
- Operates on two class and multi class problems
- Includes the following multivariate Methods: PCA, PLS-DA, ECVA, Clustering and Correlation Maps
- Directly validate your significant peaks by checking the EIC and Mass Spectra. Stay in touch
- much much more.
Directly View Significant Peaks in the Data